GATK Germline SNP INDEL V2.0
本文是Gatk Germline spns-indels Pipeline 分析遗传病(耳聋)的升级版,目的是提供开箱即用的分析流程,尽可能简化部署和迁移。
更新内容如下:
人类参考基因组以及其他引用数据库文件版本由GRCh37(hg19)升级为GRCh38(hg38)
数据注释软件annovar更换为Ensemble vep(108.2),Annovar需要商业授权,vep为apache2.0 licence,可以随意使用
Pipeline用到的软件由预先安装改为docker+conda首次使用时安装,初次运行初始化环境下载必要文件,迁移更方便
为了让pipeline能够直接运行,无需部署,这里使用docker容器保证运行环境的一致性:见文章:基于docker的生信基础环境镜像构建,这里采用的方案是带ssh服务的docker+conda环境,整个pipeline在一个通用容器中运行。
本文代码较长,可能略复杂,想直接运行的可以下载 workflow文件,导入sliverworkspace图形化生信平台直接运行,获取并查看图形化分析报告
相关代码和workflow文件可以访问笔者github项目地址
导入workflow操作

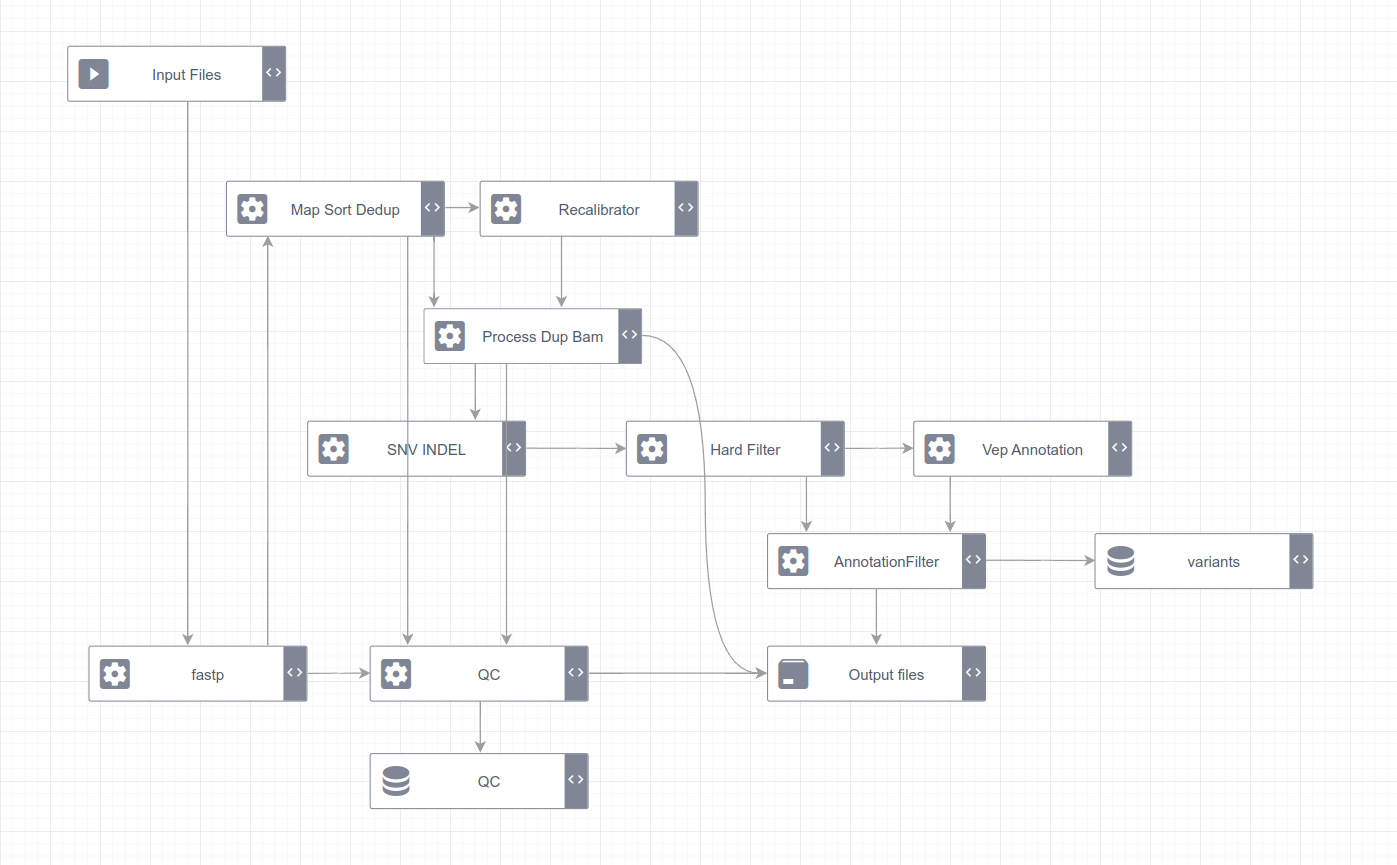
流程概览图如下
数据准备:
SRR9993255_R1.fastq.gz SRR9993255_R2.fastq.gz
SRX6732499: Targeted NGS of human: blood sample
1 ILLUMINA (Illumina HiSeq X Ten) run: 679,472 spots, 203.8M bases, 80.8Mb downloads
如果下载数据是sra,可以用NCBI官方工具sra-toolkit拆分成fastq.gz文件
fastq-dump SRR9993255 --split-3 --gzip
得到SRR9993255_R1.fastq.gz SRR9993255_R2.fastq.gz
环境搭建:
本文使用docker + conda (mamba) 作为基础分析环境,
镜像获取:docker/docker-compoes 的安装及镜像构建见:
基于docker的生信基础环境镜像构建,
bash
# 拉取docker镜像
docker pull doujiangbaozi/sliverworkspace:latest
# 查看docker 镜像
docker images基础环境配置,docker-compose.yml 配置文件,可以根据需要自行修改调整
yaml
version: "3"
services:
SarsCov2:
image: doujiangbaozi/sliverworkspace:latest
container_name: Germline
volumes:
- /media/sliver/Data/data:/opt/data:rw #挂载原始数据,放SC2目录下
- /media/sliver/Manufacture/GL2/envs:/root/mambaforge-pypy3/envs:rw #挂载envs conda环境目录
- /media/sliver/Manufacture/GL2/config:/opt/config:rw #挂载config conda配置文件目录
- /media/sliver/Manufacture/GL2/ref:/opt/ref:rw #挂载reference目录
- /media/sliver/Manufacture/GL2/result:/opt/result:rw #挂载中间文件和输出结果目录
ports:
- "9024:9024" #ssh连接端口可以按需修改
environment:
- TZ=Asia/Shanghai #设置时区
- PS=20191124 #修改默认ssh密码
- PT=9024 #修改默认ssh连接端口基础环境运行
bash
# docker-compose.yml 所在目录下运行
docker-compose up -d
# 或者
docker-compose up -d -f /路径/docker-compose.yaml
# 查看docker是否正常运行,docker-compose.yaml目录下运行
docker-compose ps
# 或者
docker psdocker 容器使用,类似于登录远程服务器
bash
# 登录docker,使用的是ssh服务,可以本地或者远程部署使用
ssh root@192.168.6.6 -p9024
# 看到如下,显示如下提示即正常登录
(base) root@SliverWorkstation:~#软件版本
| 软件名称 | 软件版本 | 备注 |
|---|---|---|
| fastqc | 0.12.1 | |
| multiqc | 1.21 | |
| bwa | 0.7.17 | |
| sambamba | 1.0.1 | static编译版本比mamba安装版本运行效率高1倍 |
| samtools | 1.16.1 | |
| bedtools | 2.30.0 | |
| fastp | 0.23.2 | |
| gatk | 4.5.0.0 | |
| ensembl-vep | 108.2 | vep软件版本 |
| version_vep_cache | 108 | vep cache 版本 |
分析结果
| 文件名 | 备注 |
|---|---|
| report.pdf | 基于报告设计器报告打印为pdf文件 |
| SRR9993255/qc/multiqc_report.html | multiqc报告文件 |
| SRR9993255/SRR9993255.result.qc.tsv | 数据质控结果 |
| SRR9993255/SRR9993255.result.variants.tsv | 致病突变结果 |
| SRR9993255/vcf/SRR9993255_filtered.vcf | 硬过滤后vcf文件 |
| SRR9993255/vcf/SRR9993255_filtered_vep.tsv | 使用vep注释后文件 |
| SRR9993255/qc/SRR9993255_depth.tsv | samtools depth获取的测序深度统计文件,用来作图显示整体测序深度 |
分析流程
变量设置:
| 变量名称 | 变量值 | 备注 |
|---|---|---|
| sn | SRR9993255 | 样本编号 |
| pn | GM02 | Pipeline代号 |
| project_bed | hg38.exon.bed | 参考基因组hg38下的全外bed文件 |
| version_reference | hg38 | 参考基因组版本hg |
| version_fastqc | 0.12.1 | |
| version_multiqc | 1.21 | |
| version_fastp | 0.23.4 | |
| version_bwa | 0.7.17 | |
| version_samtools | 1.20 | |
| version_bedtoos | 2.31.1 | |
| version_sambamba | 1.0.1 | |
| version_gatk | 4.5.0.0 | |
| version_vep | 108.2 | ensembl-vep软件版本 |
| version_vep_cache | 108 | ensembl-vep数据库版本 |
bash
#变量初始化赋值
sn=SRR9993255 #样本编号
pn=GM02 #pipeline项目编号
data=/opt/data #数据输入目录
result=/opt/result #数据输出、中间文件目录
envs=/root/mambaforge-pypy3/envs #conda安装的环境目录
threads=32 #设置可用线程数
whitelist=/opt/ref/whitelist.csv #与耳聋相关基因及突变文件
version_reference=hg38 #参考基因组版本
project_bed=hg38.exon.bed #分析流程使用bed文件,如果没有则计算一个
version_fastqc=0.12.1 #fastqc版本
version_multiqc=1.21 #multiqc版本
version_fastp=0.23.4 #fastp版本
version_bwa=0.7.18 #bwa版本
version_samtools=1.20 #samtools版本
version_bedtools=2.31.1 #bedtools版本
version_sambamba=1.0.1 #sambamba版本
version_gatk=4.5.0.0 #gatk版本
version_vep=108.2 #vep软件版本
version_vep_cache=108 #vep-cache版本1. fastp 默认参数过滤,看下原始数据质量,clean data:
bash
#conda检测环境是否存在,首次运行不存在创建该环境并安装软件
if [ ! -d "${envs}/${pn}.fastp" ]; then
echo "Creating the environment ${pn}.fastp"
mamba create -n ${pn}.fastp -y fastp=${version_fastp} fastqc=${version_fastqc} multiqc=${version_multiqc}
fi
mamba activate ${pn}.fastp
mkdir -p ${result}/${sn}/clean
mkdir -p ${result}/${sn}/qc
fastqc -t ${threads}\
${data}/${pn}/${sn}_R1.fastq.gz \
${data}/${pn}/${sn}_R2.fastq.gz \
-o ${result}/${sn}/qc &
#使用默认参数
fastp \
-w ${threads} \
-i ${data}/${pn}/${sn}_R1.fastq.gz \
-I ${data}/${pn}/${sn}_R2.fastq.gz \
-o ${result}/${sn}/clean/${sn}_fastp_R1.fastq.gz \
-O ${result}/${sn}/clean/${sn}_fastp_R2.fastq.gz \
-j ${result}/${sn}/qc/${sn}_fastp.json \
-h ${result}/${sn}/qc/${sn}_fastp.html &
wait
fastqc -t ${threads}\
${result}/${sn}/clean/${sn}_fastp_R1.fastq.gz \
${result}/${sn}/clean/${sn}_fastp_R2.fastq.gz \
-o ${result}/${sn}/qc
mamba deactivate2. 数据比对、排序、标记重复;质控
bash
#conda检测环境是否存在,首次运行不存在创建该环境并安装软件
if [ ! -d "${envs}/${pn}.align" ]; then
mamba create -n ${pn}.align -y bwa=${version_bwa} samtools=${version_samtools} bedtools=${version_bedtools}
fi
mamba activate ${pn}.align
#从github下载sambamba static 比 mamba 安装的版本速度快1倍以上.
if [ ! -f "${envs}/${pn}.align/bin/sambamba" ]; then
aria2c https://github.com/biod/sambamba/releases/download/v${version_sambamba}/sambamba-${version_sambamba}-linux-amd64-static.gz -d ${envs}/${pn}.align/bin
gzip -cdf ${envs}/${pn}.align/bin/sambamba-${version_sambamba}-linux-amd64-static.gz > ${envs}/${pn}.align/bin/sambamba
chmod a+x ${envs}/${pn}.align/bin/sambamba
fi
#github源码安装bamdst
if [ ! -f "${envs}/bamdst/bamdst" ]; then
apt-get update && apt-get install -y git make gcc zlib1g-dev
git clone https://github.com/shiquan/bamdst.git "${envs}/bamdst"
cd ${envs}/bamdst
make
cd ${result}
fi
if [ "${version_reference}" == "hg19" ]; then
echo "USE reference Version : ${version_reference}"
mkdir -p /opt/ref/hg19
#如果没有检测到参考基因组序列,则下载序列并使用bwa创建索引
if [ ! -f "/opt/ref/hg19/hg19.fasta" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/ucsc.hg19.fasta.gz -d /opt/ref/hg19 -o hg19.fasta.gz
cd /opt/ref/hg19 && gzip -d /opt/ref/hg19/hg19.fasta.gz
else
if [ -f "/opt/ref/hg19/ucsc.hg19.fasta.gz.aria2" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/ucsc.hg19.fasta.gz -c -d /opt/ref/hg19 -o hg19.fasta.gz
cd /opt/ref/hg19 && gzip -d /opt/ref/hg19/hg19.fasta.gz
fi
fi
#检查是否存在索引文件,没有则创建索引
if [ ! -f /opt/ref/hg19/hg19.fasta.amb ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.ann ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.bwt ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.pac ] ||
[ ! -f /opt/ref/hg19/hg19.fasta.sa ]; then
bwa index /opt/ref/hg19/hg19.fasta
fi
#检测samtools索引是否存在,如不存在则使用samtools创建参考基因组索引
if [ ! -f "/opt/ref/hg19/hg19.fasta.fai" ]; then
samtools faidx /opt/ref/hg19/hg19.fasta
fi
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
mkdir -p /opt/ref/hg38
if [ ! -f "/opt/ref/hg38/hg38.fasta" ]; then
aria2c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Homo_sapiens_assembly38.fasta.gz -d /opt/ref/hg38 -o hg38.fasta.gz
cd /opt/ref/hg38 && gzip -d /opt/ref/hg38/hg38.fasta.gz
elif [ -f "/opt/ref/hg38/hg38.fasta.aria2" ]; then
aria2c ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Homo_sapiens_assembly38.fasta.gz -c -d /opt/ref/hg38 -o hg38.fasta.gz
cd /opt/ref/hg38 && gzip -d /opt/ref/hg38/hg38.fasta.gz
fi
#检查是否存在索引文件,没有则创建索引
if [ ! -f /opt/ref/hg38/hg38.fasta.amb ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.ann ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.bwt ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.pac ] ||
[ ! -f /opt/ref/hg38/hg38.fasta.sa ]; then
bwa index /opt/ref/hg38/hg38.fasta
fi
#检测samtools索引是否存在,如不存在则使用samtools创建参考基因组索引
if [ ! -f "/opt/ref/hg38/hg38.fasta.fai" ]; then
samtools faidx /opt/ref/hg38/hg38.fasta
fi
fi
cd ${result}
mkdir -p ${result}/${sn}/align
bwa mem \
-t ${threads} -M \
-R "@RG\\tID:${sn}\\tLB:${sn}\\tPL:Illumina\\tPU:NextSeq550\\tSM:${sn}" \
/opt/ref/${version_reference}/${version_reference}.fasta \
${result}/${sn}/clean/${sn}_fastp_R1.fastq.gz \
${result}/${sn}/clean/${sn}_fastp_R2.fastq.gz \
| sambamba view -S -f bam -l 0 /dev/stdin \
| sambamba sort -t ${threads} -m 2G \
--tmpdir=${result}/${sn} \
-o ${result}/${sn}/align/${sn}_sorted.bam /dev/stdin
#临时增加打开文件数量,否则容易报错
ulimit -n 10240
sambamba markdup \
--tmpdir ${result}/${sn} \
-t ${threads} \
${result}/${sn}/align/${sn}_sorted.bam \
${result}/${sn}/align/${sn}_marked.bam
mv ${result}/${sn}/align/${sn}_marked.bam.bai ${result}/${sn}/align/${sn}_marked.bai
rm -f ${result}/${sn}/align/${sn}_sorted.bam
if [ "${project_bed}" == "None" ]; then
#数据文件没有附带bed文件,这里用bedtools反向计算一个
bedtools genomecov -ibam ${result}/${sn}/align/${sn}_marked.bam -bg > ${result}/${sn}/align/${sn}_bedtools.bed
bedtools merge -i ${result}/${sn}/align/${sn}_bedtools.bed > ${result}/${sn}/align/${sn}_merged.bed
samtools flagstat --threads ${threads} \
${result}/${sn}/align/${sn}_marked.bam > ${result}/${sn}/qc/${sn}_marked.flagstat &
${envs}/bamdst/bamdst \
-p ${result}/${sn}/align/${sn}_merged.bed \
-o ${result}/${sn}/qc \
--cutoffdepth 20 \
${result}/${sn}/align/${sn}_marked.bam &
else
if [ ! -f "/opt/ref/${version_reference}/${project_bed}" ]; then
mkdir -p /opt/ref/${version_reference}
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/germline/projects/${project_bed} -d /opt/ref/${version_reference}
fi
samtools flagstat --threads ${threads} \
${result}/${sn}/align/${sn}_marked.bam > ${result}/${sn}/qc/${sn}_marked.flagstat &
${envs}/bamdst/bamdst \
-p /opt/ref/${version_reference}/${project_bed} \
-o ${result}/${sn}/qc \
--cutoffdepth 20 \
${result}/${sn}/align/${sn}_marked.bam &
fi
wait
mamba deactivate3. Gatk 获取碱基质量校准table
bash
#conda检测环境是否存在,首次运行不存在创建该环境并安装软件
if [ ! -f "${envs}/${pn}.gatk/bin/gatk" ]; then
mamba create -n ${pn}.gatk -y gatk4=${version_gatk} \
r-base=3.6.2 \
r-data.table=1.12.8 \
r-dplyr=0.8.5 \
r-getopt=1.20.3 \
r-ggplot2=3.3.0 \
r-gplots=3.0.3 \
r-gsalib=2.1 \
r-optparse=1.6.4 \
r-backports=1.1.10 \
tabix \
pandas
fi
mamba activate ${pn}.gatk
if [ "${version_reference}" == hg19 ]; then
echo "USE reference Version : ${version_reference}"
#这里有个巨坑,broadinstitute ftp站点bundle目录提供的hg19版本参考文件,默认格式运行会报错,提示没有索引,使用gatk创建索引仍然报错,其实是gz格式需要使用bgzip重新压缩并且使用tabix创建索引才行
if [ ! -f "/opt/ref/hg19/dbsnp_138.hg19.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/dbsnp_138.hg19.vcf.gz -d /opt/ref/hg19
else
if [ -f "/opt/ref/hg19/dbsnp_138.hg19.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/dbsnp_138.hg19.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/hg19/dbsnp_138.hg19.vcf.gz.tbi" ]; then
gzip -k -f -d /opt/ref/hg19/dbsnp_138.hg19.vcf.gz > /opt/ref/hg19/dbsnp_138.hg19.vcf
bgzip -f --threads ${threads} /opt/ref/hg19/dbsnp_138.hg19.vcf > /opt/ref/hg19/dbsnp_138.hg19.vcf.gz
tabix -f /opt/ref/hg19/dbsnp_138.hg19.vcf.gz
fi
if [ ! -f "/opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz -d /opt/ref/hg19
else
if [ -f "/opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz.tbi" ]; then
gzip -k -f -d /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz > /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf
bgzip -f --threads ${threads} /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf > /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz
tabix -f /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz
fi
if [ ! -f "/opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz -d /opt/ref/hg19
else
if [ -f "/opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz.tbi" ]; then
gzip -k -f -d /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz > /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf
bgzip -f --threads ${threads} /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf > /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz
tabix -f /opt/ref/hg19/1000G_phase1.snps.high_confidence.hg19.sites.vcf.gz
fi
if [ ! -f "/opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/1000G_phase1.indels.hg19.sites.vcf.gz -d /opt/ref/
else
if [ -f "/opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg19/1000G_phase1.indels.hg19.sites.vcf.gz -c -d /opt/ref/hg19
fi
fi
if [ ! -f "/opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz.tbi" ]; then
gzip -k -f -d /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz > /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf
bgzip -f --threads ${threads} /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf > /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz
tabix -f /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz
fi
#创建参考序列hg19的dict字典文件
if [ ! -f "/opt/ref/hg19/hg19.dict" ]; then
gatk CreateSequenceDictionary -R /opt/ref/hg19/hg19.fasta -O /opt/ref/hg19/hg19.dict
fi
if [ "${project_bed}" == "None" ]; then
if [ ! -f "${result}/${sn}/align/${sn}_merged.bed.interval_list" ]; then
gatk BedToIntervalList \
-I ${result}/${sn}/align/${sn}_merged.bed \
-SD /opt/ref/${version_reference}/${version_reference}.dict \
-O ${result}/${sn}/align/${sn}_merged.bed.interval_list
fi
mkdir -p ${result}/${sn}/recal
gatk BaseRecalibrator \
--known-sites /opt/ref/hg19/dbsnp_138.hg19.vcf.gz \
--known-sites /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz \
--known-sites /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz \
-L ${result}/${sn}/align/${sn}_merged.bed.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/align/${sn}_marked.bam \
-O ${result}/${sn}/align/${sn}_recal.table
else
if [ ! -f "/opt/ref/${version_reference}/${project_bed}" ]; then
#根据下载的Illumina_pt2.bed 文件创建interval list文件,坐标转换,起始坐标0修改为1
#sed 's/chr//; s/\t/ /g' /opt/ref/hg19/Illumina_pt2.bed > /opt/ref/hg19/Illumina_pt2.processed.bed
mkdir -p /opt/ref/${version_reference}
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/germline/projects/${project_bed} -d /opt/ref/${version_reference}
fi
if [ ! -f "/opt/ref/${version_reference}/${project_bed}.interval_list" ]; then
gatk BedToIntervalList \
-I /opt/ref/${version_reference}/${project_bed} \
-SD /opt/ref/${version_reference}/${version_reference}.dict \
-O /opt/ref/${version_reference}/${project_bed}.interval_list
fi
mkdir -p ${result}/${sn}/recal
gatk BaseRecalibrator \
--known-sites /opt/ref/hg19/dbsnp_138.hg19.vcf.gz \
--known-sites /opt/ref/hg19/Mills_and_1000G_gold_standard.indels.hg19.sites.vcf.gz \
--known-sites /opt/ref/hg19/1000G_phase1.indels.hg19.sites.vcf.gz \
-L /opt/ref/${version_reference}/${project_bed}.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/align/${sn}_marked.bam \
-O ${result}/${sn}/align/${sn}_recal.table
fi
elif [ "${version_reference}" == "hg38" ]; then
echo "USE reference Version : ${version_reference}"
if [ ! -f "/opt/ref/hg38/dbsnp_146.hg38.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/dbsnp_146.hg38.vcf.gz -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/dbsnp_146.hg38.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/dbsnp_146.hg38.vcf.gz -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/dbsnp_146.hg38.vcf.gz.tbi" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/dbsnp_146.hg38.vcf.gz.tbi -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/dbsnp_146.hg38.vcf.gz.tbi.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/dbsnp_146.hg38.vcf.gz.tbi -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.tbi" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.tbi -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.tbi.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz.tbi -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz -c -d /opt/ref/hg38
fi
fi
if [ ! -f "/opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.tbi" ]; then
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.tbi -d /opt/ref/hg38
else
if [ -f "/opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.tbi.aria2" ]; then
echo 'download continue...'
aria2c -x 16 -s 32 ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz.tbi -c -d /opt/ref/hg38
fi
fi
#创建参考序列hg38的dict字典文件
if [ ! -f "/opt/ref/hg38/hg38.dict" ]; then
gatk CreateSequenceDictionary -R /opt/ref/hg38/hg38.fasta -O /opt/ref/hg38/hg38.dict
fi
if [ "${project_bed}" == "None" ]; then
if [ ! -f "${result}/${sn}/align/${sn}_merged.bed.interval_list" ]; then
gatk BedToIntervalList \
-I ${result}/${sn}/align/${sn}_merged.bed \
-SD /opt/ref/${version_reference}/${version_reference}.dict \
-O ${result}/${sn}/align/${sn}_merged.bed.interval_list
fi
mkdir -p ${result}/${sn}/recal
gatk BaseRecalibrator \
--known-sites /opt/ref/hg38/dbsnp_146.hg38.vcf.gz \
--known-sites /opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz \
--known-sites /opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz \
-L ${result}/${sn}/align/${sn}_merged.bed.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/align/${sn}_marked.bam \
-O ${result}/${sn}/align/${sn}_recal.table
else
if [ ! -f "/opt/ref/${version_reference}/${project_bed}.interval_list" ]; then
gatk BedToIntervalList \
-I /opt/ref/${version_reference}/${project_bed} \
-SD /opt/ref/${version_reference}/${version_reference}.dict \
-O /opt/ref/${version_reference}/${project_bed}.interval_list
fi
mkdir -p ${result}/${sn}/recal
gatk BaseRecalibrator \
--known-sites /opt/ref/hg38/dbsnp_146.hg38.vcf.gz \
--known-sites /opt/ref/hg38/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz \
--known-sites /opt/ref/hg38/1000G_phase1.snps.high_confidence.hg38.vcf.gz \
-L /opt/ref/${version_reference}/${project_bed}.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/align/${sn}_marked.bam \
-O ${result}/${sn}/align/${sn}_recal.table
fi
fi
mamba deactivate
4. Gatk 应用碱基校准、获取insert size 统计信息
bash
mamba activate ${pn}.gatk
if [ "${project_bed}" == "None" ]; then
gatk SplitIntervals \
-L ${result}/${sn}/align/${sn}_merged.bed.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-O ${result}/${sn}/interval \
--scatter-count ${scatter} &
gatk ApplyBQSR \
--bqsr-recal-file ${result}/${sn}/align/${sn}_recal.table \
-L ${result}/${sn}/align/${sn}_merged.bed.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/align/${sn}_marked.bam \
-O ${result}/${sn}/align/${sn}_bqsr.bam &
gatk CollectInsertSizeMetrics \
-I ${result}/${sn}/align/${sn}_marked.bam \
-O ${result}/${sn}/qc/${sn}_insertsize_metrics.txt \
-H ${result}/${sn}/qc/${sn}_insertsize_histogram.pdf &
else
rm -f ${result}/${sn}/interval/*.interval_list
gatk SplitIntervals \
-L /opt/ref/${version_reference}/${project_bed}.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-O ${result}/${sn}/interval \
--scatter-count ${scatter} &
gatk ApplyBQSR \
--bqsr-recal-file ${result}/${sn}/align/${sn}_recal.table \
-L /opt/ref/${version_reference}/${project_bed}.interval_list \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-I ${result}/${sn}/align/${sn}_marked.bam \
-O ${result}/${sn}/align/${sn}_bqsr.bam &
gatk CollectInsertSizeMetrics \
-I ${result}/${sn}/align/${sn}_marked.bam \
-O ${result}/${sn}/qc/${sn}_insertsize_metrics.txt \
-H ${result}/${sn}/qc/${sn}_insertsize_histogram.pdf &
fi
wait
mamba deactivate
5. Gatk HaplotypeCaller 获取snp/indel突变:
bash
mamba activate ${pn}.gatk
mkdir -p ${result}/${sn}/vcf
rm -f ${result}/${sn}/vcf/vcf-file.list
touch ${result}/${sn}/vcf/vcf-file.list
if [ "${version_reference}" == hg19 ]; then
for i in `ls ${result}/${sn}/interval/*.interval_list`;
do
rm -f ${i%.*}_bqsr.vcf.gz
gatk HaplotypeCaller \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-L $i \
-I ${result}/${sn}/align/${sn}_bqsr.bam \
-D /opt/ref/${version_reference}/dbsnp_138.hg19.vcf.gz \
-O ${i%.*}.vcf
echo ${i%.*}.vcf >> ${result}/${sn}/vcf/vcf-file.list
done
elif [ "${version_reference}" == "hg38" ]; then
for i in `ls ${result}/${sn}/interval/*.interval_list`;
do
rm -f ${i%.*}_bqsr.vcf.gz
gatk HaplotypeCaller \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-L $i \
-I ${result}/${sn}/align/${sn}_bqsr.bam \
-D /opt/ref/${version_reference}/dbsnp_146.hg38.vcf.gz \
-O ${i%.*}.vcf
echo ${i%.*}.vcf >> ${result}/${sn}/vcf/vcf-file.list
done
fi
wait
gatk MergeVcfs \
-I ${result}/${sn}/vcf/vcf-file.list \
-O ${result}/${sn}/vcf/${sn}_bqsr.vcf
mamba deactivate
6. 单个样本数据不够运行VQSR,直接执行硬过滤,过滤参考数值见#https://gatk.broadinstitute.org/hc/en-us/articles/360035532412?id=11097
bash
mamba activate ${pn}.gatk
#https://gatk.broadinstitute.org/hc/en-us/articles/360035532412?id=11097
gatk VariantFiltration \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-V ${result}/${sn}/vcf/${sn}.vcf \
-O ${result}/${sn}/vcf/${sn}_snp.vcf \
--filter-name "SNP_QD" \
--filter-expression "QD < 2.0" \
--filter-name "SNP_MQ" \
--filter-expression "MQ <40.0" \
--filter-name "SNP_FS" \
--filter-expression "FS > 60.0" \
--filter-name "SNP_SOR" \
--filter-expression "SOR > 3.0" \
--filter-name "SNP_MQRankSum" \
--filter-expression "MQRankSum < -12.5" \
--filter-name "SNP_ReadPosRankSum" \
--filter-expression "ReadPosRankSum < -8.0"
gatk VariantFiltration \
-R /opt/ref/${version_reference}/${version_reference}.fasta \
-V ${result}/${sn}/vcf/${sn}_snp.vcf \
-O ${result}/${sn}/vcf/${sn}_filtered.vcf \
--filter-name "INDEL_QD" \
--filter-expression "QD < 2.0" \
--filter-name "INDEL_FS" \
--filter-expression "FS > 200.0" \
--filter-name "INDEL_SOR" \
--filter-expression "SOR > 10.0" \
--filter-name "INDEL_ReadPosRankSum" \
--filter-expression "ReadPosRankSum < -20.0" \
--filter-name "INDEL_InbreedingCoeff" \
--filter-expression "InbreedingCoeff < -0.8"
mamba deactivate7. 使用vep注释突变结果:
bash
#conda检测环境是否存在,首次运行不存在创建该环境并安装软件
if [ ! -d "${envs}/${pn}.vep" ]; then
echo "Creating the environment ${pn}.vep"
mamba create -n ${pn}.vep -y ensembl-vep=${version_vep}
fi
mamba activate ${pn}.vep
mkdir -p /opt/result/${sn}/vcf
if [ "${version_reference}" == "hg19" ]; then
if [ ! -d "/opt/ref/vep-cache/homo_sapiens/${version_vep_cache}_GRCh37" ]; then
aria2c -x 16 -s 48 https://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/indexed_vep_cache/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz -C /opt/ref/vep-cache/
elif [ -f "/opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz.aria2" ]; then
aria2c -x 16 -s 48 https://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/indexed_vep_cache/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz -c -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh37.tar.gz -C /opt/ref/vep-cache/
fi
vep \
-i ${result}/${sn}/vcf/${sn}_filtered.vcf \
-o ${result}/${sn}/vcf/${sn}_filtered_vep.tsv \
--offline \
--cache \
--cache_version ${version_vep_cache} \
--everything \
--dir_cache /opt/ref/vep-cache \
--dir_plugins /opt/ref/vep-cache/Plugins \
--species homo_sapiens \
--assembly GRCh37 \
--hgvs \
--fasta /opt/ref/${version_reference}/${version_reference}.fasta \
--force_overwrite \
--format vcf \
--tab \
--fork ${threads} \
--offline
elif [ "${version_reference}" == "hg38" ]; then
if [ ! -d "/opt/ref/vep-cache/homo_sapiens/${version_vep_cache}_GRCh38" ]; then
aria2c -x 16 -s 48 https://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/indexed_vep_cache/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz -C /opt/ref/vep-cache/
elif [ -f "/opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz.aria2" ]; then
aria2c -x 16 -s 48 https://ftp.ensembl.org/pub/release-${version_vep_cache}/variation/indexed_vep_cache/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz -c -d /opt/ref/
tar -zxvf /opt/ref/homo_sapiens_vep_${version_vep_cache}_GRCh38.tar.gz -C /opt/ref/vep-cache/
fi
vep \
-i ${result}/${sn}/vcf/${sn}_filtered.vcf \
-o ${result}/${sn}/vcf/${sn}_filtered_vep.tsv \
--offline \
--cache \
--cache_version ${version_vep_cache} \
--everything \
--dir_cache /opt/ref/vep-cache \
--dir_plugins /opt/ref/vep-cache/Plugins \
--species homo_sapiens \
--assembly GRCh38 \
--hgvs \
--fasta /opt/ref/${version_reference}/${version_reference}.fasta \
--force_overwrite \
--format vcf \
--tab \
--fork ${threads} \
--offline
fi
mamba deactivate8. 编写脚本匹过滤突变vcf文件,vep注释后的文件,获取clivar注释致病突变
bash
mamba activate ${pn}.gatk
if [ ! -f "${envs}/GermlineVepAnnotationUtil.py" ]; then
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/GermlineVepAnnotationUtil.py -d ${envs}/
chmod a+x ${envs}/GermlineVepAnnotationUtil.py
fi
${envs}/GermlineVepAnnotationUtil.py \
-v ${result}/${sn}/vcf/${sn}_filtered.vcf \
-a ${result}/${sn}/vcf/${sn}_filtered_vep.tsv \
-o ${result}/${sn}/${sn}.result.variants.tsv
mamba deactivate
9. 编写脚本从fastp、bamdst、gatk CollectInsertSize 输出获取数据质控信息
bash
mamba activate ${pn}.gatk
if [ ! -f "${envs}/GermlineQcUtil.py" ]; then
aria2c https://raw.githubusercontent.com/doujiangbaozi/sliverworkspace-util/main/somatic/GermlineQcUtil.py -d ${envs}/
chmod a+x ${envs}/GermlineQcUtil.py
fi
python ${envs}/GermlineQcUtil.py \
--out=${result}/${sn}/${sn}.result.qc.tsv \
--sample-fastp=${result}/${sn}/qc/${sn}_fastp.json \
--sample-bamdst=${result}/${sn}/qc/coverage.report \
--sample-insertsize=${result}/${sn}/qc/${sn}_insertsize_metrics.txt
mamba deactivate
mamba activate ${pn}.fastp
multiqc ${result}/${sn}/ -f -o ${result}/${sn}/qc
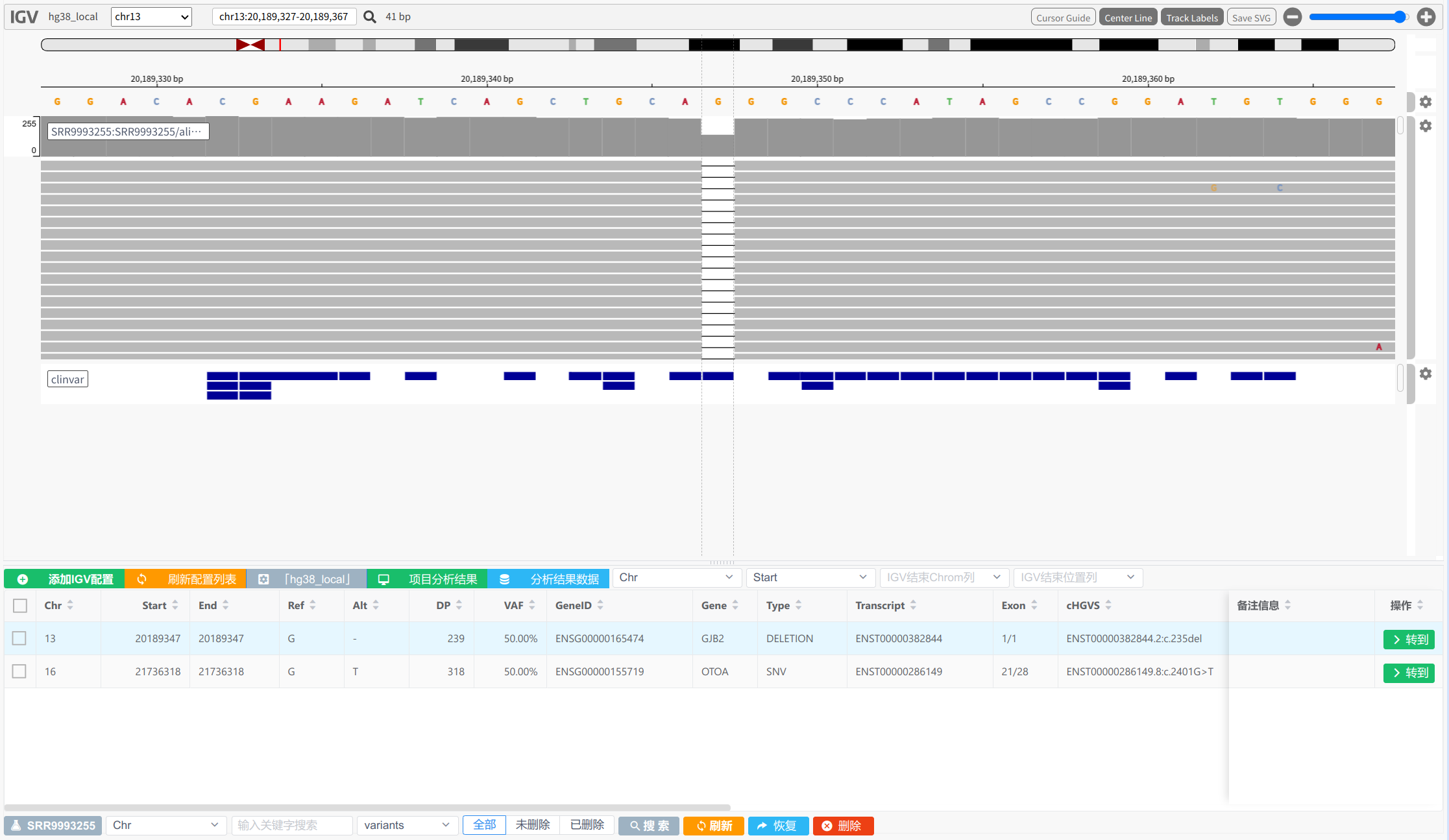
mamba deactivate结果确认,IGV bam文件和突变位置:
基于word docx模板输出的报告:

